2. Open Reading Frames (ORF)
2.1 Defintion
A reading frame is a non-overlapping set of three-nucleotide-codons (triplets)
in DNA or RNA,
which defines a gene. A codon is a set of three adjoined nucleotides that encodes
either a particular amino acid or tells the ribosome to stop translation (figure
3). Table 1 shows the 61 triplets, which code for the 20 essential amino acids,
and the 3 stop codons marked in red.
A nucleotide sequence that contains a start codon (initiation codon,
typically AUG) and a stop
codon (termination codons, UAA, UAG or UGA) is called an open
reading frame (ORF).
An ORF is hence a portion
of an organism's genome which contains a sequence of bases that could potentially encode an
individual protein. Once one knows the ORF for a gene or its mRNA, one can
translate a nucleotide sequence into its corresponding amino acid sequence,
based on the processes described in figure 3.
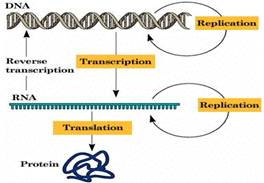 |
Figure 3: Cells whose genetic information
is stored on DNA use the two-step process to read each gene (transcription)
and produce the string of amino acids that makes up a protein (translation).
In RNA retro viruses the RNA is transcribed into a double stranded
DNA by reversed transcription before transcription and translation can take
place. The ss+ mRNA of SARS-CoV can be translated into a +strand,
which serves as template for translation and replication (see also
figure 2). |
Exercise 1: Determine the amino acid sequence of the gene
product (protein), which follows from the given segment of double stranded DNA.
The two stands are antiparallel and the nucleotide sequences are complementary.
5’GGGATCGATGCCCCTTAAAGAGTTTACATATTGCTGGAGGCGTTAACCCCGGA3’ strand 1
3’CCCTAGCTACGGGGAATTTCTCAAATGTATAACGACCTCCGCAATTGGGGCCT5’ strand 2
Indication: The DNA template strand (strand 2 above) is transcribed
by RNA polymerase into a complementary mRNA strand. In RNA-synthesis the
nucleotide chain grows from the 5’-end to the 3’end of the template and A is
translated into U, instead of T as in DNA replication. The newly synthesized
stand is antiparallel to the DNA template strand. The triplets on the mRNA strand
are then translated into an aminoacid sequence, based on the universal genetic
code (table 1).
Solution to Exercise 1:
1. Transcribe the DNA template strand into the corresponding mRNA.
5’GGGAUCGAUGCCCCUUAAAGAGUUUACAUAUUGCUGGAGGCGUUAACCCCGGA3’
2. Identify the start codon, i.e the beginning of the open reading frame.
5’GGGAUCGAUGCCCCUUAAAGAGUUUACAUAUUGCUGGAGGCGUUAACCCCGGA3’
3. Break the sequence into three-basepair-codons until you reach a stop codon.
| 5’GGGAUCG |
AUG |
CCC |
CUU |
AAA |
GAG |
UUU |
ACA |
UAU |
UGC |
UGG |
AGG |
CGU |
UAA |
CCCCGGA3’ |
4. Translate each codon into its corresponding amino acid.
| Met |
Pro |
Leu |
Lys |
Glu |
Phe |
Thr |
Tyr |
Cys |
Trp |
Arg |
Arg |
stop |
Note: ORFs are usually encountered when sifting through pieces of DNA while
trying to locate a gene.
ORFs are identified similarly in organisms which use different start-codons
based on slightly altered genetic codes.
Table 1.1 Genetic mRNA code: 61 triplets code for the 20 essential amino acids
and 3 triplets are stop signals . The direction is
5' to 3'.
|
2nd base |
| U |
C |
A |
G |
1st
base |
U |
UUU Phenylalanine (Phe/F)
UUC Phenylalanine
UUA Leucine (Leu/L)
UUG Leucine
|
UCU Serine (Ser/S)
UCC Serine
UCA Serine
UCG Serine
|
UAU Tyrosine (Tyr/Y)
UAC Tyrosine
UAA Ochre Stop
UAG Amber Stop
|
UGU (Cys/C)Cysteine
UGC (Cys/C)Cysteine
UGA Opal (Stop)
UGG (Trp/W)Tryptophan
|
| C |
CUU Leucine
CUC Leucine
CUA Leucine
CUG Leucine
|
CCU Proline (Pro/P)
CCC Proline
CCA Proline
CCG Proline
|
CAU Histidine (His/H)
CAC Histidine
CAA Glutamine (Gln/Q)
CAG Glutamine
|
CGU (Arg/R)Arginine
CGC (Arg/R)Arginine
CGA (Arg/R)Arginine
CGG (Arg/R)Arginine
|
| A |
AUU Isoleucine (Ile/I)
AUC Isoleucine
AUA Isoleucine
AUG Methionine
(Met/M) Start1
|
ACU Threonine (Thr/T)
ACC Threonine
ACA Threonine
ACG Threonine
|
AAU (Asn/N)Asparagine
AAC (Asn/N)Asparagine
AAA (Lys/K)Lysine
AAG (Lys/K)Lysine
|
AGU (Ser/S)Serine
AGC (Ser/S)Serine
AGA (Arg/R)Arginine
AGG (Arg/R)Arginine
|
| G |
GUU Valine (Val/V)
GUC Valine
GUA Valine
GUG Valine
|
GCU Alanine
(Ala/A)
GCC Alanine
GCA Alanine
GCG Alanine
|
GAU (Asp/D)Aspartic acid
GAC (Asp/D)Aspartic acid
GAA (Glu/E)Glutamic acid
GAG (Glu/E)Glutamic acid
|
GGU (Gly/G)Glycine
GGC (Gly/G)Glycine
GGA (Gly/G)Glycine
GGG (Gly/G)Glycine
|
1AUG encodes N-formylmethionine at the beginning of mRNA in Bacteria
and methionine in Archaea and Eukarya
Amino acid sequences can also be presented with the one-letter code, which
is also included in table 1.1 and again in alphabetical order in table 1.2,
e.g. the nucleotide sequence of exercise 1 (omitting stop triplet) has the following
aminoacid correspondence:
|
AUG |
CCC |
CUU |
AAA |
GAG |
UUU |
ACA |
UAU |
UGC |
UGG |
AGG |
CGU |
|
M |
P |
L |
K |
E |
F |
T |
Y |
C |
W |
R |
R |
Table 1.2. 1- and 3-letter abbreviations of amino acids
| Amino Acid |
Abbreviations |
| Alanine |
A |
Ala |
| Cysteine |
C |
Cys |
| Aspartic acid |
D |
Asp |
| Glutamate |
E |
Glu |
| Phenylalanine |
F |
Phe |
| Glycine |
G |
Gly |
| Histidine |
H |
His |
| Isoleucine |
I |
Ile |
| Lysine |
K |
Lys |
| Leucine |
L |
Leu |
| Methionine |
M |
Met |
| Asparagine |
N |
Asn |
| Proline |
P |
Pro |
| Glutamine |
Q |
Gln |
| Arginine |
R |
Arg |
| Serine |
S |
Ser |
| Threonine |
T |
Thr |
| Selenocysteine |
U |
Sec |
| Valine |
V |
Val |
| Tryptophan |
W |
Trp |
| Tyrosine |
Y |
Tyr |
2.2 Determining ORFs on the SARS genome
The complete SARS-CoV genome was published in April 2003 by a Canadian group
of
researchers. It is a single stranded RNA of 29571 bases.
Details can be found at
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&
db=nucleotide&list_uids=29826276&dopt=GenBank
The full nucleotide sequence is given for this exercise in date file sars_can.
Exercise 2: With the following steps we will define and characaterize
ORFs on the SARS genome.
Load the complete SARS-CoV sequence into the Matlab workspace
load sars_can
The detection of genes in viruses is complicated due to overlapping ORFs and
to other problems. The simplest algorithm to find ORFs in the SARS-CoV
sequence is selecting
the statistically significant ORFs among all those found by the function SEQORFs.
This function detemines the ORFs in a sequence and in its reverse complement.
If there are 3 possible reading frames in a forward mRNA strand
and considering the reverse complements will result in 6 different reading frames.
With this procedure, most of the genes can be identified. Make sure that the
m-file “seqorfs” is in your working directory.
[orf n]=seqorfs(sars_can,'MINIMUMLENGTH',1);
[orfr nr]=seqorfs(sars_can(randperm(length(sars_can))),'MINIMUMLENGTH',1);
ORFLengthr=[];
for i=1:6
ORFLengthr=[ORFLengthr; orfr(i).Length'];
end
empirical_threshold=prctile(ORFLengthr,95)
[orf n]=seqorfs(sars_can,'MINIMUMLENGTH',empirical_threshold/3);
The number “n” is the number of the ORFs found in the forward
sequence and in its reverse complement. The variable “orf” returns
a structure with start/stop positions and the length of the ORF, and it identifies
the reading frame it is in. Since we also consider the reverse complement of
the mRNA sequence of SARS-CoV, we have 6 different reading frames in our example.
To obtain for example the third one, type in
SARS_ORF=[orf(3).Start' orf(3).Stop' orf(3).Length' orf(3).Frame']
This is a matrix whose first column contains the start positions of the found
ORFs, the second column the stop positions, the third column the length of
the ORFs and the last column the corresponding frame (possible indices: {-3,-2,-1,1,2,3}).
The universal genetic code (table 1) with start codon AUG and stop codons UAA,
UAG and UGA is used by default.
Additional exercises: How many ORFs can be identified from
the entire SARS-CoV genome?
Compare your result with those given in the NCBI database. To access, type
web('http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?cmd=Retrieve&
db=Nucleotide&list_uids=30248028&dopt=GenBank&
WebEnv=0jFo_p0HwjP18h4EdA2Wei4-5l%4026414B3D6A627CB0_0063SID&WebEnvRq=1')
into the Matlab. How many ORFs code for known proteins? Can you find the ORF
that codes for the spike glycoprotein product of SARS-CoV (see figure 1)?
